AI drug discovery applies machine learning, deep learning and related computational methods to help find and optimize new medicines. It aims to reduce uncertainty across research stages by learning patterns from biological, chemical and clinical data. The result is a more data driven approach to selecting targets, designing molecules and prioritizing experiments.
Rather than replacing lab science, AI helps teams decide what to test next and what to stop early. Recent industry collaborations, such as the Nvidia and Lilly initiative detailed in this AI lab partnership transforming drug discovery, show how large scale computing and pharma expertise are converging to operationalize these methods. It also supports decisions that affect patient safety, trial success and manufacturing feasibility. Understanding how AI drug discovery works clarifies why it is becoming central to modern R and D.
What is AI Drug Discovery?
AI drug discovery is the use of algorithms to support decisions in the discovery and early development of therapeutics. It includes predictive models that estimate whether a compound will bind to a target, remain stable, dissolve, cross membranes or trigger toxicity. It also includes generative models that propose new molecular structures that satisfy multiple constraints.
This field covers small molecules, biologics, peptides and nucleic acid therapies, though methods and data types differ. Most programs combine AI with high-throughput screening, structural biology, medicinal chemistry and translational research. The strongest results appear when models are built around high quality experimental data and clear decision points.
How Does AI Drug Discovery Work?
AI drug discovery works by training models on curated datasets and using the learned relationships to predict outcomes for new targets or compounds. Inputs can include protein sequences, 3D structures, assay readouts, omics profiles, images and text from scientific literature. Outputs typically rank hypotheses so teams can allocate lab time to the most promising options.
The workflow is iterative, because each experimental round produces new labels that can improve model performance. Teams also monitor bias, uncertainty and data drift to avoid confident but wrong predictions. A practical AI system therefore includes data pipelines, model governance and a tight feedback loop with experimental science.
Typical Model Driven Loop
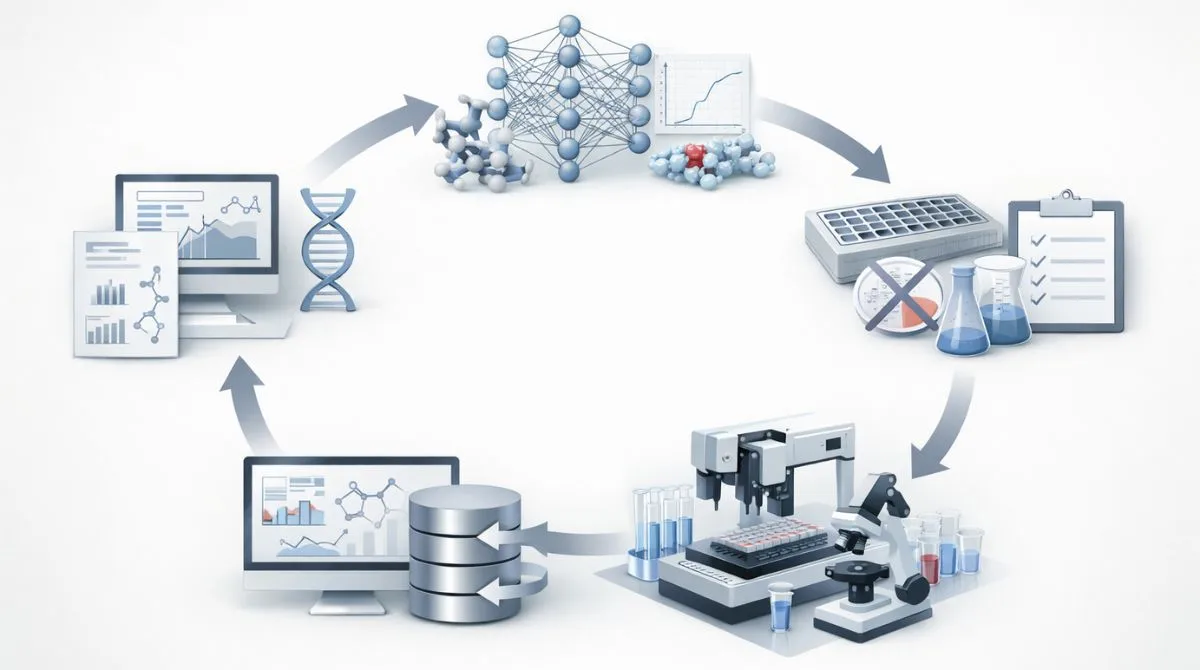
- Define the decision. Specify the exact question, such as target tractability, hit selection or off-target risk, with measurable success criteria.
- Assemble and audit data. Merge assay, chemistry and biology sources, then check label noise, missingness, and batch effects that can mislead learning.
- Engineer representations. Convert molecules and proteins into features such as fingerprints, graphs, embeddings or 3D descriptors for model input.
- Train and validate models. Use splits that reflect real deployment, apply calibration and evaluate with metrics aligned to the decision threshold.
- Prioritize experiments. Rank compounds or targets, include uncertainty estimates and choose a testing set that balances exploitation and exploration.
- Learn from new results. Feed assay outcomes back into the dataset and retrain to refine predictions and reduce error over time.
This loop only works when the experimental design and data standards are consistent across cycles. Strong documentation and reproducibility help models remain trustworthy as projects evolve.
Key AI Methods Used in Drug Discovery
Different tasks require different methods, so AI drug discovery usually combines multiple model families. The goal is not novelty but dependable performance under realistic constraints, such as limited labels and shifting chemistry series. Interpretability and uncertainty estimation are often as important as raw accuracy.
- Supervised learning. Predicts properties like potency, selectivity, solubility, metabolic stability and toxicity from labeled assay data.
- Deep learning on molecular graphs. Learns structure activity patterns directly from atoms and bonds, reducing reliance on handcrafted descriptors.
- Protein modeling and embeddings. Uses sequence or structure representations to infer binding sites, functional similarity or mutation impact.
- Generative chemistry. Proposes novel molecules conditioned on objectives such as potency and ADME while controlling novelty and synthesizability.
- Active learning. Selects the next experiments that should reduce uncertainty fastest, especially useful when assays are expensive.
- Natural language processing. Extracts relationships from papers, patents and reports to support target discovery and safety assessment.
- Causal and mechanistic hybrids. Combines statistical learning with pathway knowledge to reduce spurious correlations.
Method choice depends on data volume, assay consistency and whether the priority is ranking, classification or multi objective optimization. Teams often use ensembles and consensus scoring to improve robustness.
Where AI Fits in the Drug Discovery Pipeline?

AI can add value at nearly every stage, but the best placements are the points where decisions are frequent and costly to reverse. Early choices about targets and modalities cascade through later work, so small improvements can compound. Many organizations start with near term wins like property prediction and compound triage.
Common insertion points include target identification, hit finding, hit to lead, lead optimization, and preclinical safety assessment. AI also supports operational efficiency by improving data curation, protocol standardization and experiment tracking. When integrated well, it reduces cycles of synthesis and testing without sacrificing scientific rigor.
| Pipeline Stage | AI Contribution | Typical Output |
|---|---|---|
| Target Identification | Links genes, pathways and phenotypes using omics and literature signals | Ranked target list with evidence scores |
| Hit Discovery | Virtual screening and binding prediction to narrow chemical space | Prioritized hit set for synthesis or purchase |
| Hit To Lead | Multi-property prediction and active learning to guide analog selection | Reduced assay burden with higher quality leads |
| Lead Optimization | Generative design and property trade off modeling across potency and ADME | Optimized series proposals with risk flags |
| Preclinical Safety | Toxicity prediction, off-target profiling and metabolite risk modeling | Earlier identification of safety liabilities |
This mapping helps teams choose realistic objectives for initial deployments. As maturity grows, models can expand from single property prediction to portfolio level decision support.
Benefits of AI Drug Discovery
The main benefit is better prioritization, which reduces wasted synthesis and testing. AI can evaluate thousands to millions of candidates in silico and focus lab work on the most plausible hypotheses. It also helps programs move faster by shortening design make test cycles.
Another benefit is earlier risk detection, especially for ADMET and safety liabilities that often emerge late. Models can flag compounds with problematic reactivity, poor exposure or likely off-target activity before they consume budget. This supports a more disciplined approach to go and no-go decisions.
- Faster iteration. Shorter cycles between data generation and compound selection, which can increase the pace of medicinal chemistry.
- Expanded search space. Ability to explore chemical space beyond familiar scaffolds while still enforcing constraints like synthesizability.
- Improved multi-objective balance. Simultaneous optimization across potency, selectivity, permeability and stability instead of single-metric tuning.
- Better use of sparse data. Transfer learning and embeddings can extract signal from limited project-specific labels.
- More consistent decisions. Standardized scoring reduces variability across teams when criteria are clearly defined.
These gains depend on careful integration with experimental workflows and a willingness to measure outcomes. Clear baselines and post decision audits make it easier to prove value and refine models.
Limitations and Challenges of AI Drug Discovery
AI models inherit the weaknesses of their data, and drug discovery data is often noisy, biased and incomplete. Assays vary by lab, protocols change and negative results are underreported, which distorts learning. Without strong data governance, models can look accurate in validation and fail in real use.
Generalization is another challenge because new targets and novel chemistry can differ from training distributions. Overconfident predictions are especially dangerous when they discourage experiments that would reveal failure early. Uncertainty quantification and domain of applicability checks help, but they require discipline.
- Label noise and batch effects. Small measurement shifts can dominate the signal in structure-activity modeling.
- Limited mechanistic insight. High-performing predictors may not explain biology well, making it harder to debug errors.
- Model leakage risks. Improper data splits or duplicate compounds can inflate metrics and mislead stakeholders.
- Synthesis reality. Generative suggestions can violate practical chemistry constraints without strong filtering and retrosynthesis checks.
- Regulatory and compliance needs. Traceability, documentation and audit trails matter when models influence safety-related decisions.
These issues do not make AI ineffective, but they raise the bar for validation and monitoring. Teams that treat models as living systems usually achieve more reliable impact.
AI Drug Discovery Vs Traditional Drug Discovery

Traditional drug discovery relies heavily on sequential experimentation, expert intuition and incremental optimization guided by relatively small datasets. AI drug discovery shifts part of that burden to computational ranking and pattern learning across larger, more integrated data sources. The emphasis moves from manual hypothesis selection to systematic triage supported by predictive modeling.
In practice, both approaches converge in the lab, because biological validation remains the arbiter of truth. The difference is how decisions are made before experiments are run and how quickly teams can learn from results. Organizations that combine experienced scientists with strong modeling and data infrastructure tend to outperform either approach alone.
Traditional approaches can be more interpretable, especially when guided by known structure activity relationships and established assays. AI approaches can be more scalable and consistent, but they require investment in data quality and model governance. The best comparison is not replacement versus status quo, but improved decision quality per experiment.
Conclusion
The future of AI drug discovery depends on tighter integration between models, automation and high quality experimental data. These advances sit within a broader wave of scientific progress, where AI-driven drug discovery complements other emerging breakthroughs shaping modern biomedical research. As datasets become better curated and more representative, models should become more reliable at predicting complex outcomes like efficacy and safety. Progress will also come from hybrid approaches that combine mechanistic biology with machine learning.
Teams that succeed will focus on clear decision points, transparent evaluation and continuous monitoring of real world performance. AI will matter most where it helps scientists learn faster and fail earlier, while protecting patients through better risk assessment. With disciplined execution, AI drug discovery can make R and D more efficient and more scientifically grounded.



